Research Article
Phenotypic Plasticity and Genetic Variation of Two Wild Populations of Green tiger Shrimp (Penaeus semisulcatus - De Haan, 1844) 
 Author
Author  Correspondence author
Correspondence author
International Journal of Marine Science, 2015, Vol. 5, No. 5 doi: 10.5376/ijms.2015.05.0005
Received: 02 Nov., 2014 Accepted: 16 Dec., 2014 Published: 20 Jan., 2015
Munasinghe and Senevirathna, 2015, Phenotypic Plasticity and Genetic Variation of Two Wild Populations of Green tiger Shrimp (Penaeus semisulcatus - De Haan, 1844), International Journal of Marine Science, Vol.5, No.5: 1-8 (doi: 10.5376/ijms.2015.05.0005)
Utility of wild-caught brood stocks in shrimp farming industry is still practiced in many countries. Therefore, identification of genetic variability of geographically distributed commercially important species provide useful information that could be utilized in aquaculture programs. This study investigated the morphological and genetic variation between two wild populations of P. semisulcatus from Northern and Southern parts in Sri Lanka. Each population was consisted with 70 individuals and forty two measurements were taken according to the truss network system for morphometric study. Discriminant function analysis identified four parameters as significant contributors to discriminate two populations and they were associated with the abdominal region of P. semisulcatus. Altogether 3280 nucleotides were analyzed representing four mitochondrial and two nuclear gene regions (both coding and non-coding). Results indicated low genetic variation between two populations. Mitochondrial control gene region produced seven haplotypes which were common between two populations (nucleotide divergence ranged from 0.8% to 5.2%). Among three haplotypes resulted from Cytochrome Oxidase I gene region (COI), one haplotype was common between two populations (nucleotide divergence ranged from 0.1% to 0.3%). Other two mitochondrial gene regions (16S rRNA & 12S rRNA) and two Nuclear (18S RNA & H3) gene regions produced single shared haplotype for each region. In conclusion, results suggest the phenotypic fixation to environmental conditions indicating phenotypic plasticity in P. semisulcatus populations in northern and southern parts of the island.
1 Introduction
The green tiger shrimp, Penaeus semisulcatus shows natural distribution in Indo-West Pacific and the Mediterranean regions (Holthuis, 1980). It is one of the most important marine crustacean species subjected to intense fishery exploitation and aquaculture practices worldwide (Hulata, 2001). This species has been identified as a candidate for mass-breeding programs in some countries (Seidman and Issar, 1988; Türkmen, 2007). In Sri Lanka, green tiger shrimp fishery is mainly depend on the wild stock from all around the island. It is necessary to discriminate the unit stocks to arrive at meaningful estimates of stock bio-mass for stock assessment management. Farming of this species has not been attempted yet, but it has potential for commercial culture in Sri Lanka. The traits of the candidate should have favorable growth characteristics for seed production under farming. Environmental factors including geomorphology of the habitats play a major role in structuring populations in the wild. Also there are mechanisms that can lead to partial isolation and structuring of fish populations even in the marine environment (Hellberg, 2002). As P. semisulcatus is distributed all along the island, it is necessary to delineate the natural stocks for selection of suitable trait for farming. The study of morphometric characters and the genetic divergence pattern of wild stocks provide useful information for selection of suitable stock for farming programs.
Morphometric characters have been successfully used for taxonomic inferences. Truss network system is more reliable in collection of data in morphometric studies. Truss network measurements are a series of measurements calculated between landmarks that form a regular pattern of connected quadrilaterals or cells across the body form (Bagherian and Rahmani, 2009). Morphometry based on truss network data has been used in past studies for stock identification (Bronte and Moore, 2007; Shao et al., 2007; Bagherian and Rahmani, 2009), species discrimination (Palma and Andrade, 2002; Aktas et al., 2006), ontogeny (Hard et al., 1999; Debowski et al., 1999) and functional morphology (Dean et al., 2006). The transformed truss measurements were subjected to factor analysis and classify by cross-validation of discriminant analysis (Sen et al., 2011).
Published studies on genetic variation of P. semisulcatus populations are limited compared to other commercially important shrimp species but few studies have been documented under different aspects such as taxonomic identification (Saad et al., 2013; Khamnamtong et al., 2005) and population structure analyses (Rezvani et al., 2001; Niamaimandi et al., 2010). Utility of different gene regions in population and phylogenetic studies are important as genes evolve at different rates and thus exhibit different evolutionary patterns (Munasinghe et al. 2003). Past studies have been revealed that mitochondrial DNA (mtDNA) and nuclear DNA (nucDNA) evolve in different rates (Avise, 1994; Hartl and Clark, 1997). Therefore, pattern of genetic divergence among populations should be assessed using differently evolving gene regions for proper identification of different stocks in the wild.
Taxonomic studies on P. semisulcatus populations in Sri Lanka have not been reported so far. However, intensive exploitation of shrimps of the genus Penaeus is causing a decline in per unit landings in many areas of the seas around the world from the past (Rao et al., 1993) to the present (Mehanna et al., 2012). Therefore, identifications of stocks in different landing sites of the island may help for conservation of these species and sustainable management of the fishery industry. The aim of the present study was to investigate pheneotypic and genotypic variation of geographically separated two wild P. semisulcatus populations in Sri Lanka.
2 Materials and Methods
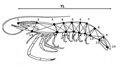
The locations of the two populations are geographically widely separated and situated at the extreme ends of the northern and southern parts of the island (Figure 1). According to the past investigations, two locations consisted with different environmental conditions (Silva et al., 2013). Samples were collected from two wild populations: Jaffna lagoon and Walawe River estuary (Figure 1). From each site, samples were collected from at least three sub sampling locations and pooled to make the final sample of the population. Approximately 70 individuals of adults were randomly selected from the collection of each population. Males and females were separated according to the structural differences of sex organs (petasma and thelycum). Forty two morphometric measurements were collected as described by Aktas et al., (2006) according to the truss network system (Figure 2). Collected data were log transformed and those skewed from normality test were omitted from further analysis.
Significant differences between sexes for all parameters within each population were determined by comparison of means using t test. Parameters that did not show significant differences between sexes for both populations were selected and thus males and females were combined (P>0.05). Finally, twenty one morphometric measurements were used to continue the analysis. To perform the multivariate analysis, all measurements were standardized using regression and residual analysis method (Thorpe, 1976). Discriminant Function Analysis (DFA) was performed to determine the most reliable morphological characters that are important in distinguishing two species. Wilks' lamda was used to test the significance of the discriminant function as a whole. Canonical scores derived from DFA analysis were plotted in two dimensional spaces for visual detection of the separation of two populations. Analysis was performed using MINITAB V. 14 (2004) and SPSS V. 20 (IBM corp. 2011).
Number of individuals analyzed from each population for each gene region is given in the Table 1. For genetic analyses individuals from each population were selected randomly. Total DNA was extracted from small muscle sample using DNEasy Extraction Kit (QIAGEN) following manufacturer's protocols. Four mtDNA gene regions and two nucDNA gene regions were partially amplified using primers published in the past studies (Table 2). Initially, amplification of PCR reactions for all six regions were conducted in 25 µL reaction mixture contained 1xTaq polymerase buffer, 1.5 mM MgCl2, 0.4 mM each dNTP, 0.2 µm of each primer, 100 ng of DNA template and 0.5 U of Taq DNA polymerase. When optimize the conditions, less primer amounts were added (0.15 µm) to 18S and H3 nuclear gene regions and additional MgCl2 was (final concentration 2 mM) added to amplify the control gene region. PCR conditions of the initial denaturation and final extension stages are common for all six regions which were used as 2 min at 95℃ and 72℃ for 7 min. respectively. PCR cycle conditions optimized for each gene region are given in the Table 2. PCR products were purified using PrepEaseTM PCR Purification 96-well Plate Kit (USB Corporation). Purified samples were sequenced at the DNA sequencing laboratory, Brigham Young University Utah, USA, using next generation sequencing method. Sequences emerged from this study were deposited in the Genbank under the given accession numbers (Table 1). Nucleotide divergence levels and phylogenetic relationships among derived sequences were analyzed using MEGA (V 5.1) program.
 Table 1 Gene regions sequenced, number of individuals used, results reveled from the of the sequencing process and Genbank Accession numbers |
3 Results
Derived discriminant function received by analyses of size-corrected data identified four morphometric parameters as significant contributors (Wilk′s Lamda: P<0.05), (Table 3). The landmarks of four morphometric parameters are 3~17, 4~15, 5~14 and 9~12. The cross validated classification showed that overall 87% were correctly classified. The box-plot graphs plotted using canonical scores derived from DFA analysis is given in the Figure 3.
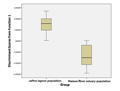 Figure 3 Graph illustrates distribution of the discriminant function scores for two populations and their separation |
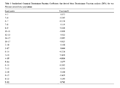 Table 3 Standardized Canonical Discriminant Function Coefficients that derived from Discriminant Function analysis (DFA) for two Penaeus semisulcatus populations |
The information of the sequences recovered for six gene regions are given in the Table 1. Mitochondrial control gene region showed high diversity level producing seven haplotypes which are common for both populations. Among three haplotypes resulted for COI gene region only one haplotype shared between two populations. Other gene regions produced single haplotype each which was common for both populations. Percentage of mean nucleotide divergence levels varied from 0.8% to 5.2% for control gene region and for COI gene region it was ranged from 0.1% to 0.3%. Neighbor Joining (NJ) trees constructed using derived sequences of mitochondrial control gene region and COI gene region are given in the Figure 4 and 5 respectively.
|
Figure 4 Neighbor Joining (NJ) tree derived from seven haplotypes of control gene region. Fenneropenaeus sp. used as an out group, Bootstrap values less than 50 are not indicated |
|
Figure 5 Neighbor Joining (NJ) tree derived from three haplotypes of COI gene region. Fenneropenaeus sp. used as an out group |
4 Discussion
Out of twenty one morphometric parameters only four parameters strongly contributed in separation of two populations. Among them diagonal length of the 3rd abdominal segment (5~14) act as the strongest predictor. Derived graph indicated the separation of two populations with slight overlapping but centroids for two populations were clearly separated (Figure 3).
This study analyzed gene regions from both mtDNA and nucDNA gene regions including coding, non-coding and hyper variable mitochondrial control gene region. Results of six gene regions indicated low genetic variation between two wild populations. High genetic variability was evident for mtDNA control gene region which produced seven haplotypes. This pattern is concordant with the past studies conducted for P. monodon populations (Chu et al., 2003; Kumar et al., 2007; You et al., 2007; Khedkar et al., 2013). The seven haplotypes separated into two distinct groups in the phylogenetic tree (Figure 4). It is evident that the availability of low genetic variation between northern and southern P. semisulcatus wild populations due to low nucleotide divergence levels among common haplotypes between two populations. The phylogenetic tree derived for haplotypes produced from COI gene region clearly separated the common haplotype (KM 528139) from other two (Figure 5). The nucleotide divergence levels derived for mtDNA control and COI gene regions were within the range when compared with the published data for other Penaeus species (Lavery et al., 2004).
According to Silva et al (2012), the Jaffna lagoon of the extreme northern and Walawe River estuary of the extreme southern regions of the Sri Lanka comprise of different environmental conditions and have been classified under two different categories. Results indicated the availability of phenotypic plasticity which means, possibility of produce different phenotypes for organisms with the same genotype under different environmental conditions (Thibert-plante and Hendry, 2011). This phenomenon is known as a common response to environmental gradients (Via et al., 1995; Agrawal, 2001) and common among marine organisms (Munday, 2013). It has been suggested that marine crustaceans those with high dispersal potential (i.e., long-lived planktonic larvae) often display phenotypic plasticity than those with low dispersal ability (Hollander, 2008). Phenotypic plasticity is also arising as heterochronic phenotypic variation, which has been reported from crustaceans especially those species undergo abbreviated larval development (Kavanagh et al., 2006; Ozawa and Ishii, 2008). It has been further suggested that the phenotypic plasticity could correspond to phenotype fixation, a kind of plasticity especially in marine environment and few studies have been reported on P. monodon in several geographic locations (West-Eberhard, 1989; Haye et al., 2010).
Although, it is evident that there is a single genetic unit in two populations, in morphological point of view, it could be suggested the availability of two morphologically different stocks of P. semisulcatus in two geographically separated locations. The occurrence of separate stocks in Sri Lankan waters may be the result of different physical and ecological condition of northern and southern sea of the island. Size and growth parameters are important characters for commercially demanded species, hence, for stock assessment it is necessary to make separate estimates for northern and southern stocks of P. semisulcatus. For aquaculture purposes, it is recommended that the breeding and growth potential of these two wild stocks should be assessed to select the suitable stock for successful hatchery production and farming.
Author contribution
Financial support for this study was from the grant received by DHNM. JDMS contributed by analyzing morphological data and DHNM collected and analyzed genetic data.
Acknowledgement
This study was partially funded by National Science Foundation Sri Lanka under the grant NSF/RG/2010/BT/02. Authors would like to thank Professor K.A. Crandall, former Head, Department of Biology, Brigham Young University, Utah, USA for providing laboratory facilities for carryout molecular analyses relevant to this study.
References
Agrawal A.A., 2001, Phenotypic plasticity in the interactions and evolution of species, Science, 294: 321-326
http://dx.doi.org/10.1126/science.1060701
Aktas M., Turan C., and Bozkurt A., 2006, Taxonomic description of three shrimp species (Melicertus kerathurus, Metapenaeus monoceros, Penaeus semisulcatus) using multivariate morphometric analysis, Journal of Animal and Veterinary Advances, 3: 172-175.
Avise J.C., 1994, Molecular Markers, Natural History and Evolution. (Chapman & Hall: New York.)
http://dx.doi.org/10.1007/978-1-4615-2381-9
Bagherian A., and Rahmani H., 2009, Morphological discrimination between two populations of shemaya, Chalcalburnus chalcoides (Actinopterygii, Cyprinidae) using a truss network, Animal Biodiversity and Conservation, 32: 1-7
Bronte C.R., and Moore S.A., 2007, Morphological Variation of Siscowet Lake Trout in Lake Superior, Transactions of the American Fisheries Society, 136: 509-517
http://dx.doi.org/10.1577/T06-098.1
Chu K.H., Li C.P., Tam Y.K., and Lavery S., 2003, Application of mitochondrial D-loop region in population genetic studies of the shrimp Penaeus, Molecular Ecology Notes, 3: 120-122
http://dx.doi.org/10.1046/j.1471-8286.2003.00376.x
Crandall K.A., and Fitzpatrick J.F., 1996, Crayfish molecular systematics: using a combination of procedures to estimate phylogeny, Systematic Biology, 45: 1-26
http://dx.doi.org/10.1093/sysbio/45.1.1
Colgan D.J., McLauchlan A., Wilson G.D.F., Livingston S.P., Edgecombe G.D., Macaranas J., Cassis G., and Gray M.R., 1998, Histone 3 and U2 snRNA DNA sequences and arthropod molecular evolution, Australian Journal of Zoology, 46: 419-437
http://dx.doi.org/10.1071/ZO98048
Dean M.N., Huber D.R., and Nance H.A., 2006, Functional morphology of jaw trabeculation in the lesser electric ray Narcine brasiliensis, with comments on the evolution of structural support in the Batoidea, Journal of Morphology, 267: 1137-1146
http://dx.doi.org/10.1002/jmor.10302
Debowski P., Robak S., and Dobosz S., 1999, Estimation of smoltification of hatchery–reared sea trout (Salmo trutta morpha trutta L.) based on body morphology, Archives of Polish Fisheries, 7: 257-266
Folmer O., Black M., Hoeh W., Lutz R., and Vrijenhoek R., 1994, DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates, Molecular Marine Biology and Biotechnology, 3: 294-299
Hard J.J., Winans G.A., and Richardson J.C., 1999, Phenotypic and genetic architecture of juvenile morphometry in chinook salmon, The Journal of Heredity, 90: 597-606
http://dx.doi.org/10.1093/jhered/90.6.597
Hartl D.L., and Clark A.G., 1997, Principles of Population Genetics, 3rd Edn. (Sinauer: Sunderland.)
Hellberg M.H., Burton R.S., Neigel J.E, and Palumbi S.R., 2002, Genetic assessment of connectivity among marine populations, Bulletin of Marine Science, 70: 273-290
Holthuis L.B., 1980, FAO Species Catalogue. Vol. 1. Shrimps and prawns of the world. An annotated catalogue of species of interest to fisheries, FAO Fisheries Synopsis 125(1): pp. 271. (FAO, Rome)
Hulata G., 2001, Genetic manipulations in aquaculture: a review of stock improvement by classical and modern technologies, Genetica, 111: 155-173
http://dx.doi.org/10.1023/A:1013776931796
Kavanagh F.A., Wilson G.D.F., and Power A.M., 2006, Heterochrony in Haplomesus (Crustacea: Isopoda: Ischnomesidae): revision of two species and description of two new species, Zootaxa, 1120: 1-33
Khedkar G.D., Reddy A.C., Ron T.B., and Haymer D., 2013, High levels of genetic diversity in Penaeus monodon populations from the east coast of India, Springer Plus, 2: 671
http://dx.doi.org/10.1186/2193-1801-2-671
Khamnamtong B., Klinbunga, S., and Menasveta P., 2005, Species Identification of Five Penaeid Shrimps Using PCR-RFLP and SSCP Analyses of 16S Ribosomal DNA, Journal of Biochemistry and Molecular Biology, 38: 491-499
http://dx.doi.org/10.5483/BMBRep.2005.38.4.491
Kumar N., Lakra W.S., Majumda K.C., Goswami M., and Ravinder K., 2007, Genetic diversity in the Indian population of Penaeus monodon (Fabricius, 1798) as revealed by mtDNA sequence analysis, Aquaculture Research, 38: 862-869
http://dx.doi.org/10.1111/j.1365-2109.2007.01740.x
Lavery S., Chan T.Y., Tam Y.K., and Chuc K.H., 2004, Phylogenetic relationships and evolutionary history of the shrimp genus Penaeus derived from mitochondrial DNA, Molecular Phylogenetics and Evolution, 31: 39–49
http://dx.doi.org/10.1016/j.ympev.2003.07.015
Medlin L., Hille J.E., Shawn S. and Sogin M.L., 1988, The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions, Gene, 71: 491-499
http://dx.doi.org/10.1016/0378-1119(88)90066-2
Mokady O., Rozenblatt S., Graur D., and Loya Y., 1994, Coral-host specificity of red sea lithophaga bivalves: interspecific and intraspecific variation in 12S mitochondrial ribosomal RNA, Molecular Marine Biology and Biotechnology, 3: 158-164
Niamaimandi N., Arshad A., Siti K.D., Ross C., Saed, and Kiabi B., 2010, Population structure of green tiger prawn, Penaeus semisulcatus (De Haan) in Bushehr waters, Persian Gulf, Iranian Journal of Fisheries Sciences, 9(2): 337-334
Ozawa H., and Ishii T., 2008, Taxonomy and sexual dimorphism of a new species of Loxoconcha (Podocopida: Ostracoda) from the Pleistocene of the Japan Sea, Zoological Journal of the Linnean Society, 153: 239-251
http://dx.doi.org/10.1111/j.1096-3642.2008.00389.x
Palma J., and Andrade J.P., 2002, Morphological study of Diplodus sargus, Diplodus puntazzo, and Lithognathus mormyrus (Sparidae) in the Eastern Atlantic and Mediterranean Sea, Fisheries Research, 57: 1–8
http://dx.doi.org/10.1016/S0165-7836(01)00335-6
Philip L.M., Rober R.W., Keyne M., John M.P., and Dustin, J.M., 2013, Predicting evolutionary responses to climate change in the sea, Ecology Letters, 16: 1488-1500
http://dx.doi.org/10.1111/ele.12185
Pilar A.H., Pilar S., Enzo A., and Elie, P., 2010, Heterochronic phenotypic plasticity with lack of genetic differentiation in the southeastern Pacific squat lobster Pleuroncodes monodon, Evolution and Development, 12(6): 628–634
Rao G.S., Subramaniam V.T., Rajamani M., Ickam P.E.S.M., and Maheswarudu G., 1993, Stock assessment of Spp. off the east coast of India, Indian Journal of Fisheries, 40(1, 2): 1-19
Rezvani G.S., Babaei S. A., and Pourkazemi M., 2001, Molecular population study on Penaeus semisulcatus from the Persian Gulf and Oman Sea using cytochrome oxidase subunit I (COI) gene by RFLP method, Iranian Journal of Fisheries Sciences, 10( 2), 15-30
Saad Y.M., Sabir J.M., and Abu Zinadah OA.H., 2013, Development of ISSR and multiplex ISSR markers for reconstructing phylogenetic relations among some shrimp species, Life Science Journal, 10: 1316 -1322
Sahar F.M., Sergiy K., Al-Kharusi L., and Al-Mamry J., 2012, Fisheries and population dynamics of the green tiger shrimp, Penaeus semisulcatus from the Arabian Sea, Oman, International Journal of Environmental Science and Engineering, 3: 33- 41
Seidman E.R., and Issar G., 1988, The culture of Penaeus semisulcatus in Israel, Journal of the World Aquaculture Society, 19: 237-247
http://dx.doi.org/10.1111/j.1749-7345.1988.tb00785.x
Sen S., Jahageerdar S., Jaiswar A.K., Chakraborty S.K., Sajina, A.M., and Dash, G.R., 2011, Stock structure analysis of Decapterus russelli (Ruppell, 1830) from east and west coast of India using truss network analyses, Fisheries Research, 112: 38-43
http://dx.doi.org/10.1016/j.fishres.2011.08.008
Shao Y., Wang J., Qiao Y., He Y., and Cao W., 2007, Morphological variability between wild populations and inbred stocks of a Chinese minnow, Gobiocypris rarus, Zoological Sciences, 24: 1094-102
http://dx.doi.org/10.2108/zsj.24.1094
Silva E.I.L, Katupotha J., Amarasinghe O., Manthrithilake H., and Ariyaratna R., 2013, Lagoons of Sri Lanka: from the origins to the present. Colombo, Sri Lanka, International Water Management Institute (IWMI).
Thorpe R.S., 1976, Biometric analysis of geographic variation and racial affinities, Biological Review, 51: 407-452
http://dx.doi.org/10.1111/j.1469-185X.1976.tb01063.x
Thibert-plante X., and Hendry A.P., 2011, The consequences of phenotypic plasticity for ecological speciation, Journal of Evolutionary Biology, 24: 326-342
http://dx.doi.org/10.1111/j.1420-9101.2010.02169.x
Türkmen G., 2007, Pond Culture of Penaeus semisulcatus and Marsupenaeus japonicus (Decapoda, Penaeidae) on the West coast of Turkey, Turkish Journal of Fisheries and Aquatic Sciences, 7: 07-11
Via S., Gomulkiewicz R., de Long G., Scheiner S.M., Shlichting C.D., and Van Tienderen P.H., 1995, Adaptive phenotypic plasticity: consensus and controversy, Trends in Ecology & Evolution, 10: 212-217
http://dx.doi.org/10.1016/S0169-5347(00)89061-8
West-Eberhard M.J., 1989, Phenotypic plasticity and the origins of diversity, Annual Review of Ecology and Systematics, 20: 249-278
http://dx.doi.org/10.1146/annurev.es.20.110189.001341
You E.M., Chiu T.S., Liu K.F., Tassanakajon A., Klinbunga A., Triwitayakorn K., and De la L.D., 2008, Microsatellite and mitochondrial haplotype diversity reveals population differentiation in the tiger shrimp (Penaeus monodon) in the Indo-Pacific region, Animal Genetic, 39: 267-277
http://dx.doi.org/10.1111/j.1365-2052.2008.01724.x
. PDF(229KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. D.H.N. Munasinghe
. J.D.M. Senevirathna
Related articles
. Morphometry
. Mitochondrial DNA
. Nuclear DNA
. Discriminant analyses
. Environment
Tools
. Email to a friend
. Post a comment