Sequence Analysis and Molecular Phylogeny of 16S rRNA Gene Fragments in Four Species of the Penaeid Shrimps from the Sudanese Red Sea 
2. School of Biological Sciences. University Sains Malaysia. Malaysia
3. Department of Zoology, Faculty of Science, University of Khartoum, Sudan
 Author
Author  Correspondence author
Correspondence author
International Journal of Marine Science, 2015, Vol. 5, No. 55 doi: 10.5376/ijms.2015.05.0055
Received: 29 Jul., 2015 Accepted: 28 Aug., 2015 Published: 12 Oct., 2015
Ibrahim M.Y., Nor M.S.A. and Abukashawa S.M.A., 2015, Sequence Analysis and Molecular Phylogeny of 16S rRNA Gene Fragments in Four Species of the Penaeid Shrimps from the Sudanese Red Sea, International Journal of Marine Science, 5(55): 1-9 (doi: 10.5376/ijms.2015.05.0055)
Penaeid shrimps are of biological and economic importance and are highly in demand for human consumption. Four species of the penaeid decapod crustaceans, Finneropenaeus indicus, Penaeus monodon, P. semisulcatus and Metapenaeus monoceros were studied. Haplotype and nucleotide diversity were combined to assess the phylogenetic relationships of the penaeid shrimp species and populations of the Finneropenaeus indicus.
Shrimp specimens were collected at different locations of Baaboud and Alkhairat aquafarms and from the wild. Phylogenetic relationships among the penaeid shrimp species and genetic diversity of F. indicus populations were assessed using partial mtDNA 16S rRNA gene (480 bp). Genetic distances among the species were done. The Genetic differentiation between F. indicus population (Baaboud –Alkhairat; Baaboud-Wild; Alkhairat- Wild) was found 0.10957, 0.12459 and 0.14817 respectively. No clear indication of differentiation between 16S rRNA tree branches of the populations of Alkhairat and Baaboud from the wild population, which may be attributed to the common collection sites of brood stocks and/or post larvae (PL’s) besides the absence of hydrological and physical barriers. M. monoceros formed a distant sister taxon to all other Penaeus species.
1 Introduction
The lack of adequate supply of animal protein, the exhaustion of livestock, the threats of malnutrition or undernourishment and the inadequacy of agricultural supplies have drawn attention to the importance of aquatic marine resources. Moreover, the demand has been extended beyond fish to include other marine invertebrates (Jackson, 1971; Ibrahim, 2001). To this attribute the production of shrimps from wild or from culture has gained a considerable contribution. Now they are considered as the most economic resource in crustacean fishery and aquafarm industry (Dall et al., 1990; Holthius, 1980; Pérez-Farfante and Kensley, 1997). Penaeid shrimps are for many centuries been considered a good source of food. (Marsden, 2008).
Approximately 125 species are known from the broad west Indo-Pacific region (Dall et al., 1990). Description and habitat of Penaeid shrimps are well documented in the Red Sea, which comprise an essential part among the fishing area 51 described by FAO (1983). Most commercially important shrimp species have complex life cycles which consist of demersal eggs that hatch into pelagic larvae which themselves go through a series of larval stages before reaching near shore nursery ground leading to development of two types of fisheries: An artisanal fishery which targets the catch of shrimps in deep narrow entrance and estuaries during migration from near shore to open sea, and the second type is: the trawling fishery in deeper water off shore (Landan, 1992). The major commercial shrimp fisheries in Sudanese Red Sea coast include F. indicus, P. monodon P. semisulcatus and M. Monoceros (Reed, 1964; El Hag, 1977) Based on their mode of life, penaeid shrimps can be grouped into wandering (e.g F. indicus) and burrowing groups (e.g P. monodon). (Dall et al., 1990).
Genetic variation is an important element with regards to the ability of the species to adapt and evolve. This measure is also used by conservation officials to plan management strategies (Schwartz et al., 2007; Reynolds et al., 2012). The spatial pattern in which genetic variation is organized within and among animal populations is referred to as genetic population structure, and in the marine environment it has been reviewed extensively by (Laikre et al., 2005) and Waples and Gaggiotti (2006).
Gene flow (gene migration) is the movement of alleles or genes within and between populations. It is influenced by a number of factors such as natural selection, genetic drift and mutation, in addition to the mobility of organisms which tend to increase the migratory potential; gene flow between two populations can also result in reducing the genetic variation between the two groups. Gene flow powerfully acts against speciation (Slatkin, 1985). Marine species with planktonic larvae are assumed demographically open, with recruitment mostly from external sources (Caley et al., 1996)
A successful aquaculture practice (after careful selection of the site) must be based on identification of the cultivable species, which is reported to be so complex in the case of shrimps owing to its phenotypic similarity and may be impossible in the absence of external characters (Palumbi and Benzie, 1991; Baldwin et al., 1998). Mitochondrial DNA (mtDNA) has been extensively used in PCR-based studies on food authenticity, population structures, phylogeography and phylogenetic relationships at different taxonomic levels (Pascoal et al., 2008).
The application of DNA markers has allowed rapid progress in aquaculture investigations of genetic variability and inbreeding, parentage assignments, species and strain identification, and the construction of high-resolution genetic linkage maps for aquaculture species (Jahmori, 2011). For molecular analysis, these markers are first amplified by PCR using conserved primers and the amplicons are sequenced. Sequencing data are then aligned and compared using appropriate bioinformatics tools. There have been considerable advances made in recent years, to provoke the ease and utilization of molecular markers. Such markers will assist in the development of the penaeid broodstock selection. One of the most efficient current methods to determine differentiation between and within species is by the use of mitochondrial DNA (mtDNA) and microsatellites (Jahromi, 2011). Molecular phylogeny of western Atlantic Farfantepenaeus and Litopenaeus shrimp based on mitochondrial 16S partial sequences was done by (Maggioni et al., 2009) .while a molecular phylogeny of the deep-sea penaeid shrimp genus Parapenaeus was studied by (Chien-Hui et al., 2015) where novel nucleotide sequence data from five different genes (COI, 16S, 12S, NaK and PEPCK) were collected to estimate phylogenetic relationships and taxonomic status amongst all but one subspecies in this genus.
On the other hand and from the commercial point of view, the black giant tiger shrimp (P. monodon) may be marketed together with the green tiger shrimp (P. semisulcatus) without any specific labeling, although recent studies have provided evidence of some divergence between both species (Pascoal et al., 2008). Also the marketing of F. indicus may be complicated to its anatomical similarities with respect to the banana shrimp F. merguiensis, which is of lower commercial value. The use of molecular tools is therefore a suitable strategy to avoid such problems.
In the present study partial 16S rRNA mtDNA gene was utilised to assess diversity and investigate the phylogenetic relationships among four shrimp species namely Finneropenaeus indicus, Penaeus monodon, P. semisulcatus and Metapenaeus monoceros.
2 Materials and Methods
2.1 Collection site and shrimps used in the study
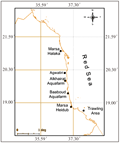
The collection sites were chosen along the Sudanese Red Sea coast which extends for 750 km in the eastern part of Africa. Two well established and easily accessible aquaculture farms, Alkhairat aquafarm and Baaboud aquafarm were chosen to represent semi- intensive and intensive mode of aquaculture respectively. Wild Mersas (Halaka in the north and Heidub in the south) and trawling areas represent wild normal shrimp fishing grounds (Figure 1).
All shrimp samples (Table 1) used in this study were brought either alive or on ice to the laboratory, sorted and identified based on morphology according to FAO (1983) and (Vine) 1986.
2.2 DNA extraction, Amplification and sequencing
Genomic DNA was extracted from 0.2 gm of frozen muscular tissue using DNeasy Tissue kit (QIAGEN, Darmstadt, Germany) following the manufacturer’s description. Extracted genomic DNA was diluted to a working concentration of 50-100 ng/µl in de-ionized water and stored in -20ºC. The partial 16S ribosomal rRNA mtDNA gene (16S rRNA gene) was amplified using universal primers 16SarF (5’-GCCTGTTTAACAAAAACAT-3’) and16SbrR (5’-CCGGTCTGAACTCAGATCATGT-3’) following (Simon et al., 1991). The PCR reagents were obtained from Vivantitis. Amplification was carried out in a total volume of 50 µL PCR mixture containing 50-100 ng of template DNA, 2.5 µL of 10x PCR buffer, 1.6 µL MgCl2, 1 µL dNTP, 2 µL of each primer, 0.2 µL of Taq DNA polymerase. Amplification was performed in a G-Storm Thermal Cycler (Gene Technologies Ltd, Braintree, Esses, CM77 6tz, UK). with a profile of pre-cycle denaturation at 94ºC for 4.30 min, followed by 30 cycles of 1.30 min at 94°C, 1 min at 51-56°C (optimizing annealing temperature), 1 min at 72°C (extension temperature), and a final extension of 5 min at 72°C. 5 µL of the PCR products were electrophoresed through a 2.5% agarose gel and stained with GelRedTM Nucleic Acid Gel Stain (10,000X in water, 0.5ml) which is proved to be more sensitive than the ethidium bromide. The amplified fragments were visualized by illumination with short wave ultraviolet light and photo documented.
Amplified DNA from individual shrimp specimens was purified by using QIAquick PCR purification kits (Qiagen, Valencia, CA) following the protocol recommended by the manufacturer. Samples of purified products were sent to Centre of Chemical Biology (CCB), USM, Malaysia for sequencing.
2.3 Sequence analysis
Mismatching alignments were checked by eye for sequence reading errors. Sequences were edited and aligned using Clustal W, Bio-Edit software (version) and CLC main workbench version 6.5 (Devereux et al., 1984). Species identifications were confirmed through Blast analysis with Genbank sequences (NCBI). And the sequence with accession number DQ149968 (Shekhar et al, 2005) was chosen as a reference because it was 100% aligned with most of the study sequences. DNA sequences of other Penaeus species were obtained from Genbank [(FJ002573 (Shekhar et al., 2008); AY751800 (Zhou and Jiang, 2004) and GU573957 (Rahnema et al., 2010)] for phylogenetic comparison. Sequence variation, and SNP events of the gene were analyzed and/or exported using Mega5.05 (Tamura et al., 2011). A neighbor-joining (NJ) tree using Kimura’s 2-parameter model (Saitou and Nei, 1987; Tamura et al., 2004) with 1000 bootstrap replicates, based on pairwise genetic distance was constructed in Mega 5.05. Dna SP version 5.10.1 (Hudson et al., 1992) was used to calculate the gene flow (Nm) and genetic differentiation (Fst).
3 Results
3.1 Amplification of 16S rRNA gene
The PCR product of approximately 480 bp from 16S rRNA gene were obtained from most of the samples, Figure 2 shows examples of amplified products of F. indicus, P. monodon, P. semisulcatus and M. monoceros.
 Figure 2 16S rRNA PCR amplified products. Lane 1: Molecular marker; Lane 2, 5: F. indicus; Lane 3: P. monodon; Lane 4: P. semisulcatus; Lane 6: M. monoceros |
3.2 Alignment and Sequences variation of the 16S rRNA gene
The partial nucleotide sequence of the gene was aligned along with the published sequence of the 16S rRNA gene region of F. indicus (gene bank: accession number DQ149968) (Figure 3). Analysis was carried on 392 bp of the amplified product. The analyzed regions of the gene were equivalent to position (44-436) in 16S rRNA reference sequence of (Shekhar et al., 2005).
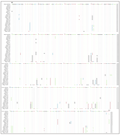 Figure 3 Sequence alignment of partial 16S rRNA gene of 4 taxa including three populations of F. indicus (Ha.I-Hap.XI), P. monodon (Hap.XII-Hap.XIII) P. semisulcatus (Hap.XIV-Hap.XVIII) and M. monoceros (Hap.XIX) from Sudanese Red Sea coast. The sequence numbered with reference to to the published sequence of the 16S rRNA gene region of F. indicus [gene bank: accession number DQ149968) Dots denote identity to the top sequence. Dashes denote gaps |
3.3 Base composition
The base composition among species (Table 2) showed slight differences between them, and the base C being the rarest (average 11.43 %) where A is the most frequent (34.14) %. (C+G) % was found to range between 32.3% in M. Monoceros to 30.8% in both of F. indicus and P. semisulcatus.
3.4 Haplotype Estimation
The alignment of 72 nucleotide sequences revealed a total of 19 haplotypes within the four penaeid species and among the three populations of F. indicus (Figure 3).
The F. indicus populations: F. indicus was found to have 11 haplotypes (from Hap.I to Hap.XI) among the entire population. The wild population had 8 haplotypes; Baaboud population had 4 haplotypes, while Alkhairat aquafarm contained only three haplotypes. It was noticed that haplotype I and IV were shared between the three locations. Haplotype I was found in 28 out of 42 (66.6%) individuals of F. indicus, hence it is the most frequent haplotype, and it was 100% aligned with the Gene bank reference (DQ149968) without any insertion or deletion, while Haplotype IV aligned with 99.7%. The wild population appeared to have more variable sites with constant nucleotide substitution at positions 50(G-C), 105(T-G), 285(C-A), 400(C-T), 410(T-C) and 438(T-G) respectively. Other minor or unique substitutions were also observed.
The P. monodon populations: Only two haplotypes (Hap.XII and XIII) were obtained from 18 individuals of P. monodon, the frequent haplotype is Hap.XIII which is found to be dominated through 15 sequences (i.e. in 83.3% of population). The P. semisulcatus populations: 5 haplotypes were encountered from P. semisulcatus.The M. monoceros: only one haplotype (Hap.XIX) was recorded for M. monoceros species.
Indel position of a codon (ATA) at position 302, 303 and 304 respectively was observed when comparing M. monoceros with the other three Penaeus species; and another indel of (A) nucleotide at position 319.
3.5 Genetic differentiation and distance
The genetic differentiation (Fst) between the three populations of F. indicus (intra-specific population) was 0.10957, 0.12459 and 0.14817. While the genetic distance between F. indicus and other species was tabulated in Table 3.
The genetic differentiation (Fst) between P. monodon populations from north (Mersa Halaka) and south (Mersa Heidub) was recorded as 0.36585.
The gene flow (Nm) is higher between the different populations in the same species as shown in Table 4. While the genetic differentiation (Fst) between F. indicus- P. monodon, F. indicus -P. semisulcatus and F. indicus-M. monoceros was found to be 0.95932, 0.94655, and 0.99016 respectively.
3.6 Phylogenetic analysis
In Figure 4 the phylogenetic tree was constructed based on the 392 bp, because not all base pairs nucleotides were sequenced completely for some specimens. The final aligned 16S rRNA sequences consisted of 392 bp, tree was generated using neighbour joining (NJ). Additional sequences (F.J002573, AY751800, Gu573957) were retrieved from gene bank to root the phylogeny. The topology of the 19 haplotypes clustered into four obvious clades with highly significant support of boot strap of values. All the eleven haplotypes of F. indicus from the three location clustered into a single clade. There is a sign of differentiation between the wild population of F. indicus into one group before it was gathered with Baaboud and Alkhairat individuals. Their gene bank reference assigned with them while F. merguiensis (F.J002573.1) form a sister clade with F. indicus clade. Haplotype IV representing at least one individual from each population grouped in a close position to F. merguiensis (F.J002573.1) considering other species on the constructed trees, it is obvious that, the 2 haplotypes of P. monodon form one clade before gathering with P. monodon (AY751800) retrieved from gene bank.
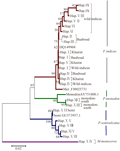 Figure 4 Neighbour-joining tree based on genetic distance analysis of 16S-rRNA sequences showing the genetic relationships of Penaeus sp. Scale shown refers to genetic distance based on nucleotide substitutions. Numbers at branching points are bootstrap support. M.monoceros appears as outgroup |
The entire five P. semisulcatus haplotypes cluster together with their gene bank reference Gu573957.1 clustered into one clade. One of them (Hap.XVI) seems to be identical with its reference. A forth distinct clade was formed by the individuals of M. monoceros with clear separation as an out group.
4 Discussion and Conclusions
In conservation genetics, understanding of the relationship between individuals is particularly important in captive breeding programmes to reduce incestuous mating in order to minimize inbreeding and the loss of genetic variation (Frankham et al., 2002). Knowledge and studies on genetics can reduce the extinction risk by helping to develop appropriate popu-lation management programmess that can minimize the risks implied through inbreeding.
The amplification of 16S rRNA products (480bp) were in the expected size range as reported by (Bouchon et al., 1994); (Shekhar et al., 2005) and Jahmori (2011). While the finfishes investigated by (Shekhar et al., 2005) have higher size of mitochondrial segment with respect to the same primers compared to crustaceans.
For all shrimp species used in the present study, there was bias towards (A+T) in the base pair composition 16S rRNA gene sequences (68.7). This is commonly the case in most penaeid species (Baldwin, et al., 1998; Maggioni et al., 2001). Furthermore, the patterns described in the present study are consistent with the description of other arthropod mtDNA sequences (Crozier et al., 1989; Garcia-Machado et al., 1993) as well as other marine crustacean mtDNA sequences (Harrison and Crespi, 1999; Meyran et al., 1997). Mitochondrial genes such as the large subunit (16S) ribosomal DNA and Cytochrome C Oxidase subunit I (COI) are popular markers used in molecular systematic studies of crustaceans at the species and population levels (e.g. Baldwin et al.,1998).
Jahmori (2011) stated that; sequences of 16S rRNA were found to be able to distinguish between 2 morphotypes of Iranian P. semisulcatus. whereas (Tsoi et al., 2005) studied two varieties of P. japonicus from South China Sea. Their results revealed that these varieties represent distinct clades, with sequence divergence of about 1% in 16S rRNA gene and 6-7% in COI gene and 16-19% in the control region, and they are separated from the other Penaeus species.
In the present study haplotype I (Hap.I) and haplotype IV (Hap.IV) were fairly common between the three populations of F. indicus from Alkhairat, Baaboud and the wild with Fst values of (0.10957, 0.12459, 0.14817) and gene flow values of Nm (4.06, 3.51, 2.87) respectively. Similar situations were encountered between the two populations of P. monodon from northern (Mersa Halaka) and southern areas (Mersa Heidub) along the Red Sea coast. To this attribute (Dall et al., 1990) and FAO (2010) described the life history of Penaeidae that comprised an offshore planktonic larval phase, an estuarine postarval and juvenile phase and an inshore adult and spawning phase. Such life cycle may allow moderate gene flow among populations, bearing in mind that F. indicus belongs to the wandering group according to its mode of life cycle and P. monodon to the burrowing group which may explain the gene flow of their each population. While shank et al. (2003) and Shanks (2009) suggested that animal species with an extended pelagic larval duration (PLD) would be more dispersive showing homogenous genetic population patterns even over very long distances.
As observed there was no clear indication of differentiation between 16SRNA tree branches of the populations of Alkhairat and Baaboud from the wild population, and support values were moderate to low. However, the mixing between population may be explained by the fact that the collection sites were common and at a narrower geographic scale structuring is not clarified as known in absence of physical and hydrological barriers, this phenomenon has been documented in many aquatic marine species, on the other hand many species lack structuring over a wide range of geographic areas (Duda and Palumbi, 1999; Avise, 2000; Benzie, 2000). However, more research is needed and more genera must be sequenced to obtain more realistic results for animals like shrimps that migrate between different habitats during their life cycle.
Authors’ Contribution
MYI participated in field sampling, Performed the technical work, analysed the results and drafted the manuscript,SAM supervised the lab work SMA supervised the research group and revised the draft manuscript. All authors read and approved the final manuscript.
Acknowledgement
Thanks are due Mr. Sami Saeid Ali for his support in collection of the samples from the aquafarms (Alkhairat and Baaboud) and to Mr. Ezzaldein, and Mr.Shakir( Red Sea University) and Miss Amal(Khartoum University) for their help during field and lab work in Sudan. Special thanks to Dr. Mashair Sir Elkhatim for her support and help during lab work at Malaysia, and to U of K colleagues during analysis. The research was sponsored by Red Sea University. Sudan.
References
Avise, J.C., 2000, Phylogeography: The history and formation of species. Harvard University Press. Cambridge, Massachusetts.
Baldwin, J.D., Bass, A.L., Bowen, B.W. and Clark, W.H., Jr.,1998, Molecular phylogeny and biogeography of the marine shrimp Penaeus. Mol. Phylogenet. Evol. 10:399-407
http://dx.doi.org/10.1006/mpev.1998.0537
Benzie, J. A. H., 2000, Population genetic structure of penaeid prawns. Aquaculture Research 31:95-119
http://dx.doi.org/10.1046/j.1365-2109.2000.00412.x
Bouchon, D.; Grosset; C. S and Raimond, R., 1994, Mitochondrial DNA variation and markers of species identity in two penaeid shrimp species: Penaeus monodon (Fabricius) and P. japonicus (Bate). Aquaculture 127:131-144
http://dx.doi.org/10.1016/0044-8486(94)90420-0
Caley, M.J.; Carr, M.H., Hixon; M.A., Hughes, T.P.; Jones, G.P. and Menge, B.A., 1996, Recruitment and the local dynamics of open marine populations. Annu. Rev. Ecol. Syst. 27:477-500
http://dx.doi.org/10.1146/annurev.ecolsys.27.1.477
Chien-Hui Y., Zhongli Sha, Tin-Yam C. and Ruiyu L., 2015, Molecular phylogeny of the deep-sea penaeid shrimp genus Parapenaeus (Crustacea: Decapoda: Dendrobranchiata). Zoologica Scripta. Vol. 44-3: 312-323
Crozier, R.H.; Crozier, Y.C.; and Mackinlay, A.G.,1989, The CO-I and CO-II region of the honeybee mitochondrial DNA: Evidence for variation in insect mitochondrial evolutionary rates, Mol. Bio. Evol., 6:399-411
Dall, W.; Hill, B.J.; Rothlisberg, P.C. and Staples D.J., 1990, The biology of Penaeidae. In: Blaxter JHS, Southward AJ (eds) Advances in marine biology Vol. 27. Academic Press, New York, USA.
Devereux, J.; Haeberli, P. and Smithies, O.,1984, A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12:387-395
http://dx.doi.org/10.1093/nar/12.1Part1.387
Duda, J.r., and Palumbi, S. R., 1999, Population structure of black tiger prawn, Penaeus monodon, among western Indian Ocean and western Pacific population. Marine Biology,134: 705-710
http://dx.doi.org/10.1007/s002270050586
El Hag, E. A.,1977, Ecological and some biological aspects of the Sudanese estuarine prawns (PENAEIDAE). M. Sc. Thesis. Department of Zoology, Faculty of Science, University of Khartoum, Sudan. 61pp.
FAO.,1983, FAO species identification sheet, fishing area 51(W. Indian ocean).
Frankham, R.; Ballou, J. D. and Briscoe, D. A., 2002, Introduction to conservation genetics. Cambridge Univ. Press, New York.
http://dx.doi.org/10.1017/CBO9780511808999
Garcia-Machado, E.; Dennebouy, N.; Suarez, M. O.; Mounolou, J. C., and Monnerot, M. ,1993, Mitochondrial 16s-rRNAgene of two species of shrimps: Sequence variability and secondary structure. Crustaceana 65(3):279-286
http://dx.doi.org/10.1163/156854093X00711
Harrison, M. K., and Crespi, B. J., 1999, Phylogenetics of cancer crabs (Crustacea: Decapoda: Brachyura). Mol. Phylogenet. Evol. 12(2):186-199
http://dx.doi.org/10.1006/mpev.1998.0608
Holthius, L.B., 1980, FAO species catalogue. Volume 1. Shrimps and prawns of the world. An annotated catalogue of species of interest to fisheries. FAO Fisheries Synopsis, 125(1):1-271
Hudson, R. R., Slatkin, M and Madisson W. P., 1992, Estimation of levels of gene flow from DNA sequence data, Genetics 132:583-589
Ibrahim, Mona Y., 2000, Some Studies on some Holothurians species along the Sudanese Red Sea coast. M. Sc. Thesis. Department of Zoology. Faculty of Sciences. U of K Sudan.
Jackson, R.I., 1971, The importance of fish inspection in the national utilization of fishery resources in fish inspection and quality control. R. Krewer. Ed. Whitefars Press Ltd London
Jahromi, S.T., 2011, Morphological and molecular analysis of Penaeus semisulcatus from the Persian Gulf and Oman Sea. Ph. D thesis. University Science Malaysia (USM), Malaysia.
Laikre, L., Palm, S., and Ryman, N., 2005, Genetic population structure of
fishes: Implications for Coastal Zone Management. AMBIO: A Journal of
the Human Environment 34, 111-119
Landan, M., 1992, Introduction to Aquaculture. John Wiley & Sons Inc.
Maggioni, R.; Rogers, A. D.; Maclean, N. and D’Incao, F., 2001, Molecular phylogeny of Western Atlantic Farfantepenaeus and Litopenaeus shrimp based on mitochondrial 16Spartial sequences. Mol. Phylogenet. Evol. 18(1):66-73
http://dx.doi.org/10.1006/mpev.2000.0866
Marsden, G., 2008, Factors affecting productive performance of the prawn (P. monodon) Ph. D
thesis. School of Natural Resources Sciences, Queensland University of Technology.
Meyran, J.C.; Monnerot, M. and Taberlet, P., 1997, Taxonomic status and phylogenetic relationships of some species of the genus Gammarus (Crustacea, Amphipoda) deduced from mitochondrial DNA sequence. Mol Phylogenet. Evol. 8:1-10
http://dx.doi.org/10.1006/mpev.1996.0399
Palumbi, S.R. and Benzie, J., 1991, Large mitochondrial DNA differences between morphologically similar Penaeus shrimp. Mol. Mar. Biol. Biotechnol. 1:27-34
Pascoal, A; Barros-Velzquez, J.; Cepeda, A.; Gallardo, J. M and Calo-Mata, P., 2008, A polymerase chain reaction–restriction fragment length polymorphism method based on the analysis of a 16S rRNA/tRNAVal mitochondrial region for species identification of commercial penaeid shrimps (Crustacea: Decapoda: Penaeoidea) of food interest, Electrophoresis 29: 499-509
http://dx.doi.org/10.1002/elps.200700406
Pérez-Farfante, I. and Kensley, B., 1997, Penaeoid and sergestoid shrimps and prawns of the world. Keys and diagnoses for the families and genera. Paris: editions du Muséum National d’Histoire Naturelle.
Rahnema, R., Hosseini, S. J, Yavari, V., Zolghrnein, H., Qasemi, S. A. and Metinfar, A., 2010, Penaeus semisulcatus 16Sribosomal RNA gene, partial sequence; Direct submission
Reed, W., 1964, Red Sea Fisheries of Sudan. Government Printing Press. Khartoum. 116pp.
Reynolds, L.K., McGlathery, K.J., and Waycott, M., 2012, Genetic Diversity Enhances
Restoration Success by Augmentin Ecosystem Services. PLoS ONE 7, e38397
http://dx.doi.org/10.1371/journal.pone.0038397
Robson, C. M., 1979, Purification and properties of the digestive amylase of Asellus aquaticus (Crustacea: isopoda). Comp. Biochem. Physiol., 62B:501
http://dx.doi.org/10.1016/0305-0491(79)90124-x
Saitou, N. and Nei, M., 1987, The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425
Schwartz, M.K., Luikart, G., and Waples, R.S., 2007, Genetic monitoring as a promising tool for conservation and management. TRENDS in Ecology and Evolution 22
http://dx.doi.org/10.1016/j.tree.2006.08.009
Shekhar, M. S.; Gopikrishna, G. and Azad, I.S., 2005, PCR-RFLP analysis of 12S and 16S mitochondrial rRNA genes from brackish water finfish and shellfish species. Asian Fisheries Science 18: 39-48
Shanks, A.L., 2009, Pelagic larval duration and dispersal distance revisited. Biology Bulletin 216, 373-385
Shanks, A.L., Grantham, B.A. and Carr, M.H., 2003, Propagule dispersal distance and the size and spacing of marine reserves. Ecological Applications 13. S159-S169
http://dx.doi.org/10.1890/1051-0761(2003)013[0159:PDDATS]2.0.CO;2
Shekhar, M. S.; Gopikrishna, G. and Azad, I.S., 2005, PCR-RFLP analysis of 12Sand 16Smitochondrial rRNA genes from brackish water finfish and shellfish species. Asian Fisheries Science 18: 39-48
Shekhar, M.S., Gopikrishna, G., Vinaya K., Gopal,C., Pillai, S. M.and Ravichandran, P., 2008, Species Identification of white shrimp by PCR-RFLP and sequence analysis of mitochondrial genes .Unpublished.
Simon, C.; Franke, A. and Martin, A., 1991, The polymerase chain reaction: DNA extraction and amplification. In: Hewitt GM, Johnson Slatkin AWB and Young JPW (eds), Molecular Techniques in Taxonomy. Springer-Verlag, Berlin, pp 329-355
http://dx.doi.org/10.1007/978-3-642-83962-7_22
Slatkin, M., 1985, Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16:393-430
http://dx.doi.org/10.1146/annurev.es.16.110185.002141
Stark, J.R. and Walker, R.S. (1983). Carbohydrate digestion in Pecten maximus. Comp. Biochem. Physiol., 73B:173
http://dx.doi.org/10.1016/0305-0491(83)90190-6
Tamura, K.; Nei, M. and Kumar, S., 2004, Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences (USA) 101:11030-11035
http://dx.doi.org/10.1073/pnas.0404206101
Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M. and Kumar, S., 2011, MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, doi : 10.1093/molbev/msr121
http://dx.doi.org/10.1093/molbev/msr121
Tsoi, K.H.; Wang, Z.Y. and Chu, K.H., 2005, Genetic divergence between two morphologically similar varieties of the kuruma shrimp Penaeus japonicus. Mar Biol 147:367-379
http://dx.doi.org/10.1007/s00227-005-1585-x
Vine. P., 1986, Red Sea invertebrates. Immel publishing, London 224pp.
Waples, R.S., and Gaggiotti, O., 2006, What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molecular Ecology 15:1419-1439
http://dx.doi.org/10.1111/j.1365-294X.2006.02890.x
Wigglesworth, J. M. and Griffith, D. R. W., 1994, Carbohydrate digestion in Penaeus monodon. Mari. Biol., 120:571
http://dx.doi.org/10.1007/BF00350077
Zhou, F. and Jiang, S., 2004, Genetic diversity of Penaeus monodon in Sanya of Hainan using mtDNA 16S rRNA gene sequence analysis. Unpublished.
. PDF(661KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Ibrahim M.Y.
. Nor M.S.A.
. Abukashawa S.M.A.
Related articles
. Sequence analysis
. F. indicus
. P. monodon
. P. semisulcatus
. Ribosomal RNA gene (rRNA)
. Sudanese Red Sea
Tools
. Email to a friend
. Post a comment
.jpg)


